-
Notifications
You must be signed in to change notification settings - Fork 176
Description
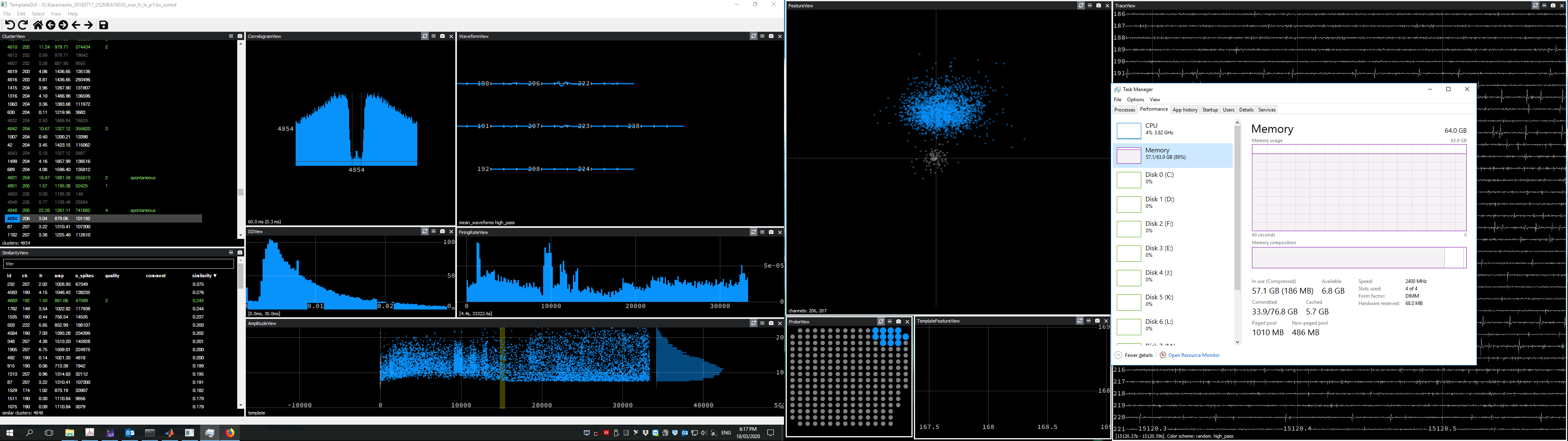
phy2 is giving us some trouble with large datasets. We have many-hour-long recordings from the retina with 252 electrodes. Besides from the binary files being large at around 150-200GB, some experiments come with millions of spikes detected by Kilosort (100-300 million). Hence, many of the .npy files written by Kilosort are very big. For an example experiment, the size of pc_features.npy is ~43 GB and template_features is ~14 GB. We use external SSDs for both the binary files and .npy files.
After starting phy, we naturally start looking at some clusters and performing splits and merges. The RAM used by phy slowly starts ramping up, till it almost maxes out. Then, instead of phy becoming faster over time because of caching, it actually starts becoming slower or unresponsive, and the whole pc is also getting slower. We noticed that memory tends to rise more after performing a lot of splits (lasso tool in the Feature or AmplitudeView).
The only solution to the problem is to periodically restart phy. I don't know how easy it will be to reproduce this problem, but do you have any ideas on how to overcome it?
In the past, we were using phy1 for similar datasets, but this particular problem was never there. Another thing I've noticed is that the hard-drive caching (.phy folder) is much smaller in size with phy2 (2-4 GB), while with phy1 the .phy folder could sometimes exceed 100 GB.