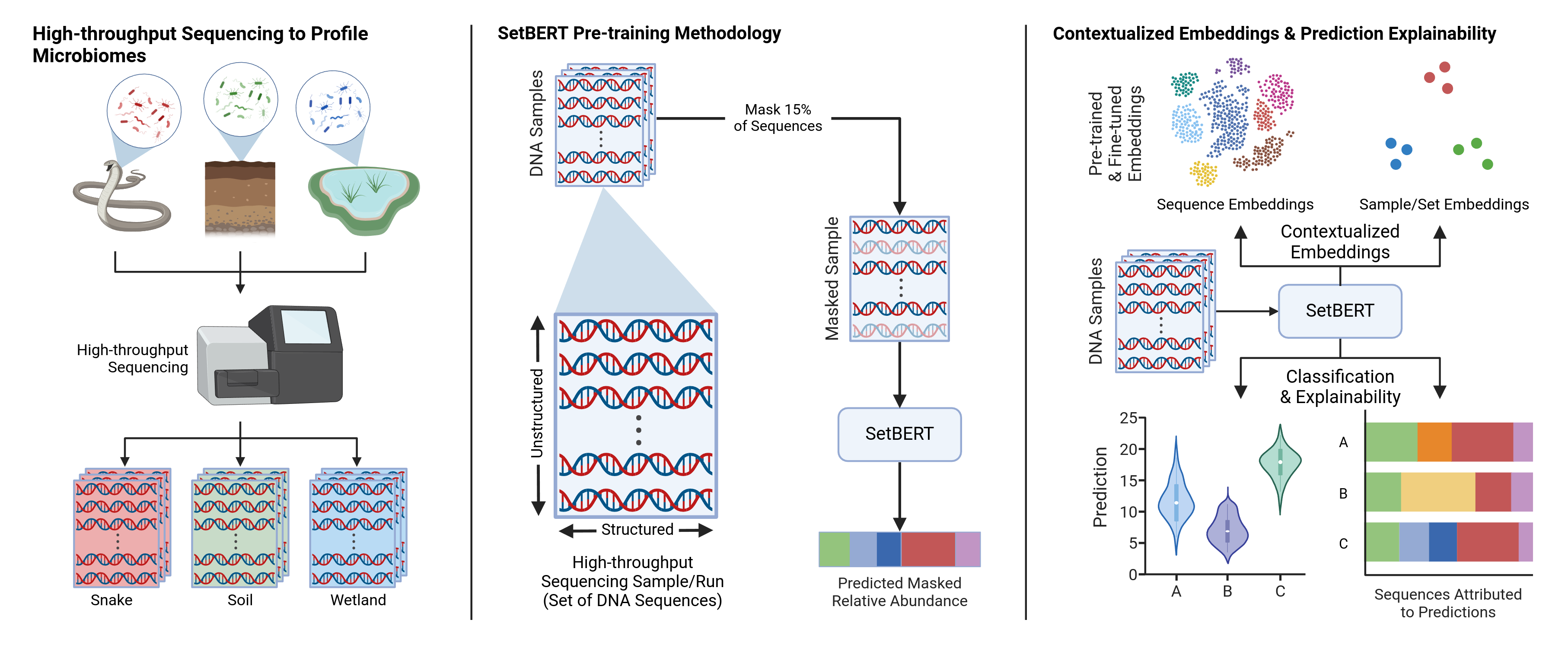
The official code repository for SetBERT: SetBERT: the deep learning platform for contextualized embeddings and explainable predictions from high-throughput sequencing
Installation from PyPI:
pip install dbtk-setbertDownload SetBERT pre-trained on the Qiita 16S platform (see Available Models for other options):
from setbert import SetBert
# Download the model
model = SetBert.from_pretrained("sirdavidludwig/setbert", revision="qiita-16s")
# Get the tokenizer
tokenizer = model.sequence_encoder.tokenizerExample sample embedding
import torch
# Input sample
sequences = [
"ACTGCAG",
"TGACGTA",
"ATGACGA"
]
# Tokenize sequences in the sample
sequence_tokens = torch.stack([torch.tensor(tokenizer(s)) for s in sequences])
# Compute embeddings
output = model(sequence_tokens)
# Sample level representation
sample_embedding = output["class"]
# Contextualized sequence representations
sequence_embeddings = output["sequences"]| Model Revision | Platform | Pre-training Dataset Description |
|---|---|---|
qiita-16s |
16S Amplicon | ~280k 16S amplicon samples from the Qiita platform |
SetBERT embeds the DNA sequences in chunks using activation checkpointing. This chunk size is specified
by the sequence_encoder_chunk_size parameter in the SetBert.Config class and adjusted freely at any point.
# Set chunk size
model.config.sequence_encoder_chunk_size = 256 # default
# Remove chunking and embed all sequences in parallel
model.config.sequence_encoder_chunk_size = Nonegit clone https://github.com/DLii-Research/setbert
pip install -e ./setbert@article{ludwig_setbert_2025,
title = {{SetBERT}: the deep learning platform for contextualized embeddings and explainable predictions from high-throughput sequencing},
volume = {41},
issn = {1367-4811},
doi = {10.1093/bioinformatics/btaf370},
number = {7},
journal = {Bioinformatics},
author = {Ludwig, II, David W and Guptil, Christopher and Alexander, Nicholas R and Zhalnina, Kateryna and Wipf, Edi M -L and Khasanova, Albina and Barber, Nicholas A and Swingley, Wesley and Walker, Donald M and Phillips, Joshua L},
month = jul,
year = {2025},
}The original source code used to produce the models and experiments for the manuscript are available in the bioinformatics branch of this repository.