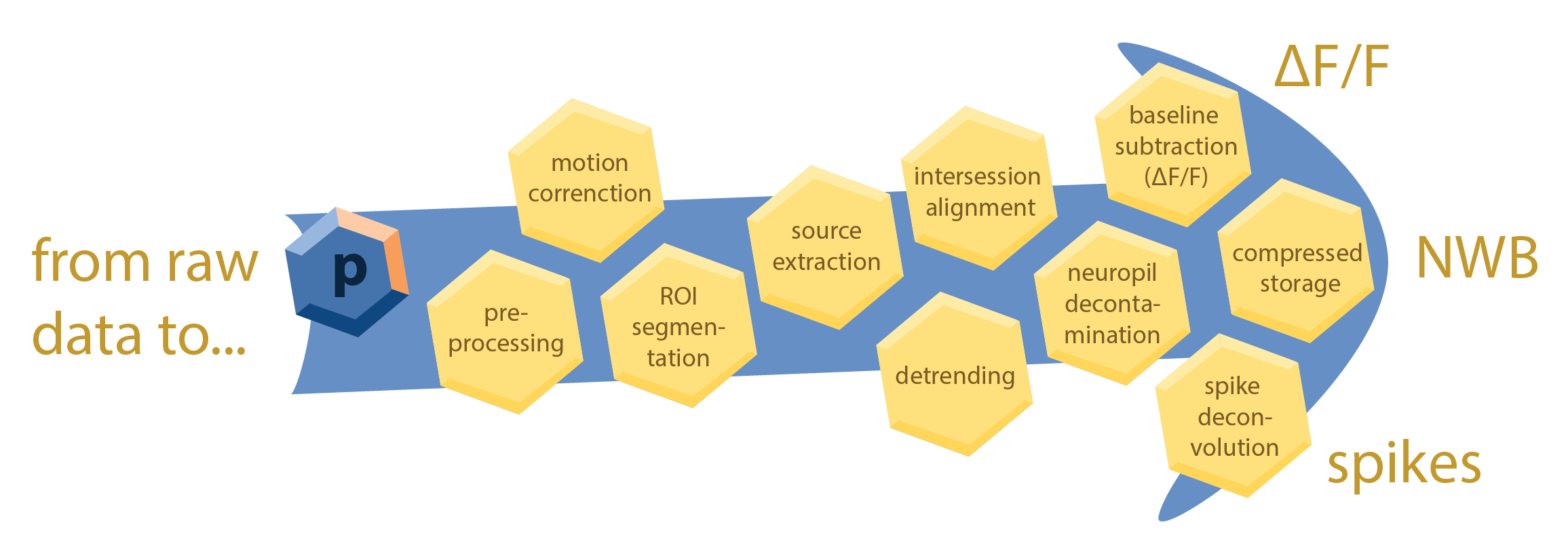
photon-mosaic is a Snakemake-based toolkit for processing multiphoton datasets. It orchestrates a curated collection of algorithms to transform your raw data (e.g., TIFF files) into analysis-ready outputs, such as ΔF/F traces, NWB files, or inferred spikes.
Each analysis step is integrated into an automated workflow, allowing you to chain preprocessing, registration, signal extraction, and post-processing steps into a single, reproducible pipeline. The design prioritizes usability for labs that process many imaging sessions and need to scale across an HPC cluster.
This is made possible by Snakemake, a workflow management system that provides a powerful and flexible framework for defining and executing complex data processing pipelines. Snakemake automatically builds a directed acyclic graph (DAG) of all the steps in your analysis, ensuring that each step is executed in the correct order and that intermediate results are cached to avoid redundant computations. photon-mosaic also includes a SLURM executor plugin for Snakemake to seamlessly scale your analysis across an HPC cluster. To ensure consistency and reproducibility, photon-mosaic writes processed data according to the NeuroBlueprint data standard for organizing and storing multiphoton imaging data.
The goal of photon-mosaic is to provide a modular, extensible, and user-friendly framework for multiphoton data analysis that can be easily adapted to different experimental designs and analysis requirements. For each processing step, we aim to vet and integrate the best available open-source tools, providing sensible defaults tailored to the specific data type and experimental modality.
- Preprocessing: derotation and contrast enhancement (see
photon_mosaic/preprocessing). - Registration & source extraction using Suite2p.
- Cell detection / anatomical ROI extraction using Cellpose (v3 or v4, including Cellpose-SAM).
- Registration using NoRMCorre for non-rigid motion correction.
- ROI matching using ROICat for inter-session / inter-plane ROI matching.
- Neuropil subtraction / decontamination: methods from the AllenSDK and AST-model.
- Spike deconvolution: OASIS and CASCADE.
See issues on GitHub and the project board for more details and participate in planning. Please refer to our guidelines to understand how to contribute.
Photon-mosaic requires Python 3.11 or 3.12.
conda create -n photon-mosaic python=3.12
conda activate photon-mosaic
pip install photon-mosaicTo install with developer tools (e.g., testing and linting):
pip install 'photon-mosaic[dev]'